XML Exporter (for PubMed and ISI)
DOS Batch program to export XML and SGML files to PubMed and ISI. Located in c:\scielo\xml_scielo.
For PubMed, there are two types of files:
-
- Journal data:
- http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=helplinkout.section.files.Resource_File#files.Resource_File_Format. Sent once or every time the journal data was changed.
-
- Articles data:
- http://www.ncbi.nlm.nih.gov/entrez/query/static/spec.html Sent one XML file for each issue.
Configuration
If it is the first installation, you have some procedures to execute.
There is a file in c:\scielo\xml_scielo\config.example. You have to copy and rename it to config.
Configure the files:
- PubMed\doi_conf.txt
- PubMed\config\config.seq
- PubMed\journals\journals.seq
File doi_conf.txt
It contains the data of the Publisher and the prefix given by CrossRef, according to the agreement signed by CrossRef and the SciELO of each country. IF YOUR SCIELO DOES NOT HAVE IT. SO THIS FILE MUST BE EMPTY.
INSTITUTION SPACE E-MAIL SPACE PREFIX
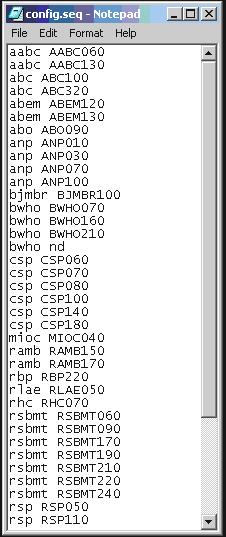
File config\config.seq
The file configconfig.seq is to inform to the program which articles or text must not be sent to PubMed, because some kind of documents are not accepted, and it is know by the section in the table of contents.
Acronym space sectionId
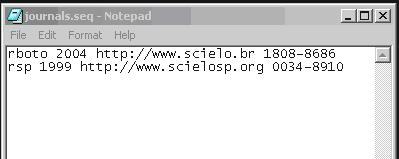
File journals\journals.seq
This file journals\journals.seq contains data used to generate XML file of the journal: journals_acronimo.xml.
This is the first XML file which must be sent to PubMed in order to register the journal. Read more: http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=helplinkout.section.files.Resource_File#files.Resource_File_Format.
Its format is:
ACRONYM SPACE FIRST_YEAR_IN_PubMed SPACE SCIELO_URL SPACE ISSN
One line for each journal.
Executing
It has to be executed using the command line in DOS.
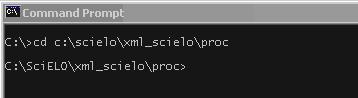
Go to the folder where this program is installed. E.g.: c:\scielo\xml_scielo\proc.
In proc you will find three scripts:
-
GenerateXML_all.bat: generates at the same time ISI and PubMed
-
GenerateXML_ISI.bat: generates SGML to ISI
-
GenerateXML_PubMed.bat: generates XML to PubMed
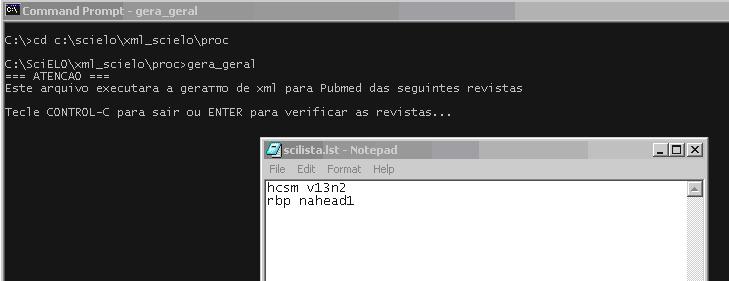
Provide a list similar to scilist, to execute any of them.
The program will open the scilist file and you have to check it, and include or remove lines, according to what you want to generate.
To generate also the XML file which contains journal data, journals_<acronimo>.xml, add one more parameter “YES”:
To generate ONLY the XML file which contains journal data, journals_<acronimo>.xml, the second parameter must be “NONE” and third one must be YES.
To generate XML file of ahead articles, use as:
- fourth parameter: the start date
- fifth parameter: the end date
The program will generate the XML file for articles which has ahpdate (publication date of ahead) between 20140100 and 20140228.
The name of the XML file will be hcsm2014nahead20140100-20140228.xml.
SciELO XML to PubMed XML
Program to export XML to PubMed, according to http://www.ncbi.nlm.nih.gov/books/NBK3828/, using SciELO XML (SciELO Publishing Schema).
How to execute
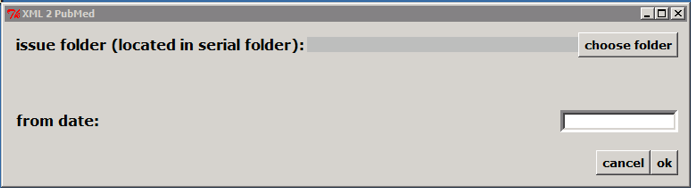
Double clicking on c:\scielo\bin\xml\xml_pubmed.py
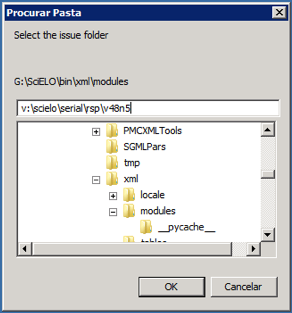
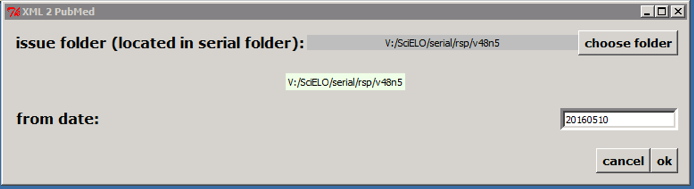
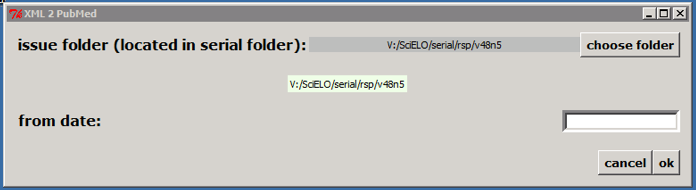
Select the issue folder
Only if issue is published on batches, such aop or rolling pass, you should inform from date to generate XML for the article published from this date to the current date.
Then click on OK button.
According to the example, the program will create the file: v:\scielo\serial\rsp\v48n5\PubMed\rsp-v48n5-20160510-20160523.xml, containing articles which have epub date between 20160510 and the current date.
If it is not an issue published on batches, click on OK button. According to the example, the program will create the file: v:\scielo\serial\rsp\v48n5\PubMed\rsp-v48n5.xml.
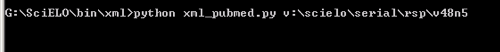
Or execute it on a terminal:
Optionally informing the from date


No comments to display
No comments to display